DNA Racial Classifications Refuted!
by Yair Davidiy
Case Study: "The Cohen Gene"
DNA Theory divides the world's populations according to haplogroups. Every change in each haplogroup is attributed to a single ancestor. The following article proves that the said changes may occur repeatedly at different times and encompass whole segments of otherwise unrelated people in specific areas. This effectively nullifies to a great degree the applicability of DNA studies to historical research regarding the movements and interrelationships of different peoples!
To demonstrate our point in the article below we concentrated on The Cohen Modal Haplotype (CMH) popularly known as "The Cohen Gene".

Brit-Am
Research
and Discovery
Facing A Revision of DNA Theory
DNA Racial Classifications Refuted!
by Yair Davidiy on behalf of Brit-Am
And in response to interest expressed by Brit-Am supporters.
Elul 5766.
Abstract: Scientists classify different groups of mankind according to differences in their DNA structure. Examination of Y DNA determines racial male ancestry and mtDNA defines ancestral groupings along the female lines. These classifications are based on the assumption that hereditary changes in DNA are one-time only events experienced in one individual who transmits the new differentiation to his descendants. In other words, it is being said that each change has only one human ancestor. Case study evidence shows that the said assumption is incorrect. The same changes occur at more than one time and in more than one subject. They do in fact appear across whole spectrums of population. This is exemplified by the Cohen Modal Haplotype (CMH) which amongst Jews emerged spontaneously within the seed of Aaron while independently also appearing in other peoples, as we show. These facts limit the applicability of DNA racial classification in the historical sense for large groups of people. A new formulation is needed and a rethinking of the applicability of DNA to historical studies of ethnic permutation.
DNA (Deoxyribonucleic acid) is a kind of helix (double twisted) string of molecules containing genetic instructions. DNA comes packed into chromosomes which are associated groups of DNA segments known as genes. Females have 23 XX chromosomes while males have 22 XX chromosomes and one XY chromosome. The Y in the XY chromosome in theory is passed from father to son and is not influenced by the mother. By tracing unique characteristics in the Y chromosome we may trace male ancestry.
In simple terms, Chromosomes are located in the nucleus of all cells of the human body. The nucleus is surrounded by miniscule agents called mitochondria that convert organic materials into energy to keep the cells going. Mitochondria have their own DNA which until recently was considered to pass only from female to female, i.e. a male receives his mitochondria from his mother but does not pass it on whereas a female both receives it from her mother and passes it onto her children. By examining changes in mitochondria we may trace female ancestors (woman after woman) back through the generations. This is mtDNA.
Conventional DNA Theory as presented to the public more or less says the following:
In the case of Y DNA: A mutation occurs in a portion of the DNA of an individual's Y Chromosome that will differentiate him from everybody else. He will pass his now unique DNA onto his male descendants and they to theirs until further down the line another change occurs in the DNA of one other descendant and so the process will begin again. The result is that we may use these differences to distinguish between major groups and subgroups and so classify humanity into different Y DNA Haplogroups.
The same concept exists for mtDNA.
The changes are presented as occuring one time only, as being
"unique event polymorphisms (UEP)".
The Implications
By stressing the one-time only aspect, UEP (Unique Event Polymorphisms), humanity may be stratifed into different groupings and sub-groupings the relationships of one to another being quite clear or at least may be presented as such.
DNA Y Haplogroups
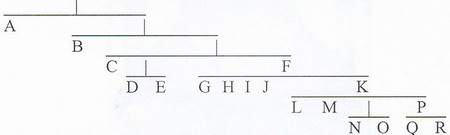
If you look at the above diagram and if necessary read our brief explanation in the article on Y DNA you will see that according to present-day understanding of Y DNA development Y haplogroups form the following clusters:
(i) (A, B, D, E) Sub-Saharan Africans, North African Berbers, Southern Mediterraneans, Japanese and Tibetans belong in one group .
(ii) (F, G, H, I, J) Caucasian Mountain Peoples, European Balkan Groups, Jews and Middle East Peoples, along with many Indians, Sardinians, Bosnians, Germans, and Scandinavians.
(iii) (K, L, M, P, N, O, Q, R) This interrelated group comprises
Indonesians, South-east Asians, Chinese, Koreans, Fins, Slavs, Western Celts, Basques, and Amerindians.
There may be factors we are unaware of. One could even try and align the above "clusters" with the Bible by perhaps equating (i) with Ham, (ii) with Shem, and (iii) with Japhet.
This is however a bit difficult for us (especially in light of our Brit-Am Biblical and Historical researches) to agree with DNA "science" in accepting the above divisions. It seems unlikely that most of the Irish and inhabitants of the British Isles (R1b) are closer to the Chinese in origin than they are to the Jews, Scandinavians, and Germans. We also find it a bit strange to view a majority portion of the Japanese as closer to the Zulus than they are to the Chinese and Koreans.
If this really was what Science says we would be faced with the choice of either accepting it or putting it to one side in the expectation that eventually new findings would effect a revision that was more acceptable to our "gut" instinct.
This however is NOT what DNA research actually says but rather how it has been presented.
An Alternate Proposal
In the DNA literature one often comes across assumptions or facts that can only be explained by the presence of what we will call REP (Repeat Event Polymorphisms).
By REP we mean that instead of UEP (Unique Event Polymorphisms) being the rule they are the exception.
A DNA sequence reflects the state of a living organism. It has an inbuilt resilience allowing it to switch from one state to another under given circumstance.
The organism will react according to its inbuilt coding when encountering external stimuli.
Not only will one single organism react in the said way but large numbers when affected at the same time by the same circumstances.
According to this we would propose, for instance, that regardless of the haplogroups that originally reached a certain area external influences at certain stages cause them all to convert in a specific direction, e.g. J, G, and I (or some such) may anciently have come to the British Isles but in the course of time they all changed due to the environment or what not to R1b.
Sound fantastic?
It could be, yet archaeological DNA studies do indicate a much great diversity in the West than currently exists. Hg (Haplogroup) K for instance is an offshoot of the (F, G, H, I, J) group yet considered the forefather of
(K, L, M, P, N, O, Q, R). Hg K apart from Finland is rare now in Europe but in the ancient Basque area reached 16% and so did hg J which is not present there now. Similar phenomenon have been reported from much of Europe.
It may also be that changes do take place but only in certain directions and only under set conditions with a certain haplogroup being able to change only along limited parameters, etc.
Nevertheless our proposal does appear to be within the boundaries of accepted scientific possibility and in accordance with known phenomenon.
As an example of this ability within DNA Coding to change in more than one location
(in REP, Repeat Event Polymorphisms) we have chosen to examine a case that has received much publicity and that is frequently held up as an example of the efficacy of DNA studies. Our example is an examination of the Cohen Modal Haplotype (CMH).
The Cohen Modal Haplotype (CMH)
Haplogroup J is a subdivision of F and is part of a grouping that includes haplogroups G, H, I, J and is prominent in the Caucasian Mountains, in the Balkans (hg G), amongst Middle East Peoples (hg J), some Indians (hg H), a portion of the Scandinavians, Germans, Bosnians, and Sardinians (hg I).
About 40% of Jews are haplogroup J.
The CMH is a subgroup of haplogroup J.
Even though J accounts only for 40% of the Jews as a whole it reaches more than 80% amongst the Cohanim.
The Jewish population is divided into Cohens, Levites, and ordinary Israelites who amongst the Jews are mostly from Judah and Benjamin.
<<Within Jewish groups, membership in three male castes (Cohen, Levi, and Israelite) is determined by paternal descent (Behar et al. 2003).>>
The Cohens are descended from Aaron the brother of Moses. They are the Priestly Caste. They were originally part of Levi but today they are perhaps more numerous than the Levites are. They also have an entirely different genetic structure from the Levites.
There are "Ashkenazic" (European) Jews and "Sephardic" (Mediterranean and Eastern) Jews.
Haplogroup J consists of an ancestral form (J*) and two subgroups J1 and J2.
<<Among Ashkenazim, J2 occurs among 23.2% of the population, while Sephardim have 28.6% (Semino et al. 2004). While these percentages are nearly identical to Iraqi (22.4%) and Lebanese (25%) groups, they are also comparable to Greek (20.6%), Georgian (26.7%), Albanian (19.6%), Italian (20-29%), and to a lesser extent, French Basque (13.6%) populations (Semino et al. 2004).>>
<<J1 is the only haplogroup that researchers consider Semitic in origin because it is restricted almost completely to Middle Eastern populations, with a very low frequency in Italy and Greece as well (Semino et al. 2004).>>
<<According to Behar (2003), the Cohanim possess an unusually high frequency of haplogroup J in general, reported to comprise nearly 87% of the total Cohanim results. Among the Sephardim, the frequency of 75% is also notably high (Behar 2003). Given the high frequency of haplogroup J among Ashkenazi Cohanim, it appears that J2 may be only slightly less common than J1, perhaps indicating multiple J lineages among the priestly Cohanim dating back to the ancient Israelite kingdom.>>
The CMH is a subgroup of haplogroup J.
Although you can have the CMH in either J1 or J2, it is the genetic signature in J1 that is considered the Jewish priestly signature.
<<Overall, J1 constitutes 14.6% of the Ashkenazim results and 11.9% of the Sephardic results (Semino et al. 2004). >>
48% of Ashkenazi Cohanim and 58% of Sephardic Cohanim have the J1 Cohen Modal Haplotype (Skorecki et al. 1997).
To sum up, about 40% of the the Jews are hg J (both J1 and J2) whereas for Cohanim the figure is ca. 80% out of which about half (ca. 40%) have the CMH whereas for Jews who are not Cohens only ca. 3% have the CMH. Most Cohens who are J1 are also CMH wheas most non-Cohen J1 Jews are not.
Does CMH Define the Cohens?
No.
About half the Cohens do not have CMH and CMH is also found in other peoples.
CMH Amongst Non-Jews
Avshalom Zoossmann-Diskin
"Are today's Jewish priests descended from the old ones?"
HOMO: Journal of Comparative Human Biology - Zeitschrift fuer
vergleichende Biologie des Menschen
51:2-3 (Urban & Fischer Verlag, 2000): 156-162.
Zoossmann explains that:
* The Cohen modal haplotype is the most common haplotype among
Southern Italians*1, Central Italians*2, Hungarians*3, and Iraqi
Kurds*4, and is also found among many Armenians*5 and South African Lembas*6. .
cf. J.E. Elkins
<<The bulk of the CMHg chromosomes were observed in J1 (53.0%) and J2 (43.2%), with a small portion falling outside of haplogroup J (3.8%). Members of the CMHg were observed throughout the world, with significant frequencies in various Arab populations: Oman (20.1%), Iraq (15.2%), Palestine (9.5%).>> An Updated World-Wide Characterization of the Cohen Modal Haplotype. J.E. Ekins et al.
Is the CMH the result of a one-time only mutation (UEP, Unique Event Polymorphism) as conventionally explained?
The CMH CANNOT BE the result of a single event but can only be explained as a recurring phenomenon effecting a change that after taking place is passed on by hereditry. It mainly takes place within haplogroup J but in 3% of the cases has also been recorded outside of it.
To explain how this could be so we must clarify the difference between haplogroup and haplotype.
Haplogroups are determined by SNP (pronounced "snip") while haplotypes are defined by STP.
<<In examining Y chromosomal diversity in this review, two types of data are considered: Single Nucleotide Polymorphisms (SNPs), and Short Tandem Repeat Loci (STRs). STR markers are characterized by mutation rates much higher than those seen with SNPs. SNPs, on the other hand, are derived from rare nucleotide changes along the Y chromosome, so-called unique event polymorphisms (UEP). These UEPs represent a single historical mutational event, occurring only once in the course of human evolution. UEPs have been given a unified nomenclature system by the Y Chromosome Consortium (2002), resulting in the identification of each UEP with a particular haplogroup.>>.
In principle STPs have a faster rate of change than SNPs which are considered more stable. Different STPs may therefore be found on the one SNP. Different STPs define Haplotypes whereas differences in the SNP define the Haplogroup. Haplotypes should therefore be considered the same as subdivisions of Haplogroups. The problem is that the same Haplotype is found frequently in more than one Haplogroup. Haplotypes are transmitted by hereditry but they are an example of a typical REP (Repeat Event Polymorphism) that can occur more than once and in more than one context. They are an example of Identical Genetic Change that occurs in more than one place in more than one subject amd then are inherited and can be used to trace ancestry. Since the same changes however occur in other bodies their applicability is limited. If we knew what influences the occurence of the changes we could use the hereditry aspects. If for instance we were aware that groups within a certain area at a certain time were liable to have undergone the said changes when we later find these changes in other places we could assume that their ancestors had formerly sojourned in the affected regions at the time in question. This however would not be enough to determine hereditry though it could lead in that direction.
The CMH is a haplotype that occurs in J1, J2, and elsewhere:
<<The bulk of the CMHgchromosomes were observed in J1 (53.0%) and J2 (43.2%), with a small portion falling outside of haplogroup J (3.8%)>>
An Updated World-Wide Characterization of the Cohen Modal Haplotype.
J.E. Ekins et al.
Placing the occurrence of CMH outside of J to the side for the moment and concentrating on its occurrence only in J1 and J2 Giacomi made a proposal that superficially could explain the occurrence of CMH in both J1 and J2.
<<The CMH is considered the putative ancestral haplotype of haplogroup J1 (Coffman: Di Giacomo et al. 2004).>>
Giacomi says that the CMH gave rise to J1. If this were so it would explain perhaps how CMH could be found in both J1 and J2. The explanation could be explained as saying that originally there was J2. CMH emerged as a haplotype in J2. From the CMH variety of J2 emerged J1. Originally all J1 would have had CMH but something happened, J1 changed and produced varieties mostly without CMH. Nevertheless CMH remained as a sub-group in J1 and also as a sub-group in J2 from which it had originally emerged. This scenario is a bit far-fetched and even Giacomi probably would not agree with it. It is however the only way the presence of CMH in both J1 and J2 can be explained while preserving the principle of it being the result of a one-time only event. The problem however is that CMH is considered YOUNGER than both J2 and J1 and therefore could not have fathered "J1".
From: Dienekes Pontikos <dienekes.pontikos@gmail.com>
The CMH can arise on a J1 and a J2 background independently. In
addition to the CMH, there are numerous other haplotypes shared by
both J1 and J2, and there is no reason to believe that the CMH in
particular was the ancestral haplotype of haplogroup J. Actually, it
was almost certainly not, because the CMH has a long DYS388-16, and
ancestral J almost certainly had a short DYS388.
What Does the CMH Really Tell Us?
Question: We have seen from the above that CMH does occur at a much higher frequency amongst Cohans than amongst others. You have shown that CMH is also found in non-Jewish people. You have also shown that not all bearers of CMH are necessarily related to each other in anyway.
What is then is significance of CMH concerning Jews or people who think they may be Jewish?
Answer: CMH is more likely to appear in hg J (especially J1?) than in other haplogroups. Once CMH appears it is transmitted by hereditry. Since Cohens are the most likely to have CMH when CMH is found amongst Jews there is a higher than average chance that the Jew is a Cohen.
Assuming that our figures are correct if ca. 6% of Jews are Cohens and half of them are CMH then ca.3% of the Jews are CMH Cohens. We also understand that 3% of Jews who are not Cohens are CMH. If therefore we come across a Jew with CMH there is a probability factor of one out of two that he is a Cohan against a chance of 3 out of 100 when we do not know if he is CMH or not. In other words, half the Jews with CMH are Cohens and a Jew with CMH is ca.17 times more likely to be a Cohen than a Jew without CMH.
We can go further than this. Half the Cohens have CMH. Jews who are Cohens are aware of the fact because they have a family tradition that they are Cohens. There are however numerous reasons why this tradition would not always have been kept. We can exprect to find Jews who are Cohens and not aware of it. The number of Jews who are not Cohens (as far as they know) with CMH equals the numbers of Cohens who have it. It could be that many of these non-Cohen CMH Jews actually had a Cohan ancestor. Within Jews we can assume that CMH is particular to Cohens or almost so. Since twice as many Cohens as "Israelite-Jews" are hg J and half of these are CMH we are justified in regarding the CMH in Cohens as emerging amongst the Cohanim some time after the designation of their forefather Aaron the first Cohen! This explains why about half the Cohens are not CMH.
CMH is a haplotype that emerges spontaneously and after emerging remains to be transmitted by hereditry. This means that all Cohens who have CMH could be related to each other but CMH in non-Jewish peoples in most cases could be an independent phenomenon.
When CMH is found amongst non-Jews it does not indicate that the person has Jewish ancestry. If however additional evidence is available the presence of CMH may have some supportive value.
By analogy if ca. 14% of Irishmen have red hair versus only ca.7% for the non-Irish inhabitants of Glasgow, Scotland, and 10%(?) of Glasgow is Irish when we find a red-haired person in Glasgow there is (all other factors being the same) one chance in five (instead of one chance in ten) that the person is an Irishmen. This does not mean that all red-haired people are related to the Irish since not all red-hairs are kinfolk to each other even though red-hair is often an inherited trait.
Conclusion:
We see from the above that DNA changes take place spontaneously and repeatedly under certain conditions.
DNA haplogroups therefore have only LIMITED APPLICABILITY in determining common ancestry. They do however have some pertinence since once the DNA changes take place they are transmitted by hereditry. DNA may be able to tell us which groups of people were in the same areas at the same time and subject to the same influences even though they were not necessarily related to each other. More study on these matters is required.
The Cohen Modal Haplotype (CMH) demonstrates the use of DNA by possibly helping us to trace who is a Cohen within the Jewish Community. On the other hand the CMH also exemplfies the limitations of DNA studies when we find it appearing independently amongst other peoples and with no ancestral connection to Israel.
See also:
Biblical Proof
Brit-Am DNA
THE PHYSICAL ANTHROPOLOGY OF
THE HEBREW PEOPLES
Queries about Race
Queries about DNA
mtDNA
Y DNA
R1b The Western Japhet?? or not?
haplogroup I
Sources
Most of the above quotations UNLESS OTHERWISE INDICATED are from:
A MOSAIC OF PEOPLE: THE JEWISH STORY AND A REASSESSMENT OF THE DNA EVIDENCE
http://www.jogg.info/11/coffman.htm
Ellen Levy-Coffman
Di Giacomo F, Luca F, Popa LO, Akar N, Anagnou N, Banyko J, Brdicka R, Barbujani G, Papola F, Ciavarella G, Cucci F, Di Stasi L, Gavrila L, Kerimova MG, Kovatchev D, Kozlov AI, Loutradis A, Mandarino V, C. Mammi C, Michalodimitrakis EN, Paoli G, Pappa KI, Pedicini G, Terrenato I, Tofanelli S, Malaspina P, Novelletto A (2004) Y chromosomal haplogroup J as a signature of the post-neolithic colonization of Europe. Hum Genet 115:357-71
Main Page
Offerings and Publications
Return to Question and Answer
Table of Contents


